model = Model6SV(wavelength_array=np.arange(350, 1000, 10))
model.show("matplotlib")100%|██████████| 65/65 [00:09<00:00, 7.19it/s]This module can be imported using: from openhsi.atmos import *
This module contains a 6SV1.1 Atmospheric Correction class that computes the pixel radiance given some parameters and a servable interactive SNR widget. 6SV is an open souce radiative transfer code that predicts the solar spectrum received by a sensor facing the Earth. Since it is written in Fortran, we use a Python wrapper called Py6S to expose the functionality and that means we are limited to using 6SV1.1 rather than the newer 6SV2.1.
Additionally, an interactive widget to compute the reflectance using Empirical Line Calibration (ELC) is provided.
Find the closest station at http://weather.uwyo.edu/upperair/sounding.html to use the atmospheric sounding on the day. find the station number and region code (for example, south pacific is pac and new zealand is nz) Default is Willis Island in Queensland, Australia. https://py6s.readthedocs.io/en/latest/helpers.html#importing-atmospheric-profiles-from-radiosonde-data
Py6S states that custom aerosol profiles can be ingested but this is broken and there is no easy fix. Don’t waste a day trying…
The Model6SV class is callable on itself. Each time the class is initialised or called, the radiance will be recalculated. The results can be viewed using Model6SV.show which returns a figure object created using the bokeh or matplotlib backends.
Model6SV.show (plot_lib:str='bokeh')
plot the calculated radiance
| Type | Default | Details | |
|---|---|---|---|
| plot_lib | str | bokeh | choose between the “bokeh” or “matplotlib” plotting backend |
| Returns | Curve | output plot |
Py6S provides a method to loop over an array and return the 6SV result. However, it does not have a progress bar. For something that can take several minutes, not knowing how long it will take is genuinely frustrating. Therefore, we provide a modified version of SixSHelpers.Wavelengths.run_wavelengths called Model6SV.run_wavelengths that has a progress bar.
To use 6SV, Py6S will write input files and parse the output files. All this I/O is slow but we got to deal with it. Py6S is clever about it though and uses threading to do some compute while waiting for files - this ends up calling the 6SV executable in parallel. The modified version follows the same method except with the addition of a callback to update the progress bar when interations are done.
Model6SV.run_wavelengths (wavelengths:<built-infunctionarray>, n_threads:int=None)
Modified version of SixSHelpers.Wavelengths.run_wavelengths that has a progress bar. This implementation uses threading (through Python’s multiprocessing API).
If you used conda to install Py6S, then the Fortran code is compiled for you with the executable path set in the install. If installing from pip, then you’ll need to follow additional instructions here and quote the path to the executable.
100%|██████████| 65/65 [00:09<00:00, 7.19it/s]Now that the at sensor radiance is calculated, the apparent reflectance can be found by dividing your measured radiance by it.
In order to use Empirical Line Calibration (ELC), there needs to be targets with known spectra in the scene such as calibration targets. A spectral library is what we need to feed into the ELC algorithm.
remap (x, in_min, in_max, out_min, out_max)
convert x from between input range (in_min,in_max) to output range (out_min,out_max).
SpectralLibrary (speclib_path:str=None)
Manages all the spectral library and interpolations as necessary.
SpectralLibrary.dump (save_path:str=None)
Dump the spectral library to file. If no save_path provided, overwrite the original file.
The spectral library should be a pandas DataFrame with the first column called wavelength and the rest of the columns labelled by the name. The data should be in reflectance (for example, measured using an ASD spectrometer or similiar).
| wavelength | spectralon | gray_small | green_small | red_small | cyan_small | gray | charcoal | blue | blue_take_off | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 350 | 1.004860 | 0.088191 | 0.071970 | 0.098128 | 0.085883 | 0.101887 | 0.058318 | 0.098823 | 0.092749 |
| 1 | 351 | 1.004398 | 0.089546 | 0.072579 | 0.098610 | 0.085919 | 0.102933 | 0.059781 | 0.100108 | 0.093942 |
| 2 | 352 | 1.004371 | 0.089546 | 0.071572 | 0.096913 | 0.084025 | 0.102238 | 0.059273 | 0.099544 | 0.093412 |
| 3 | 353 | 1.004620 | 0.087913 | 0.069308 | 0.093765 | 0.080976 | 0.099957 | 0.057630 | 0.097805 | 0.091682 |
| 4 | 354 | 1.004296 | 0.085593 | 0.067664 | 0.091687 | 0.079096 | 0.098291 | 0.057134 | 0.097217 | 0.090731 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2146 | 2496 | 1.006842 | 0.239516 | 0.333679 | 0.338185 | 0.334990 | 0.274833 | 0.062169 | 0.322644 | 0.273633 |
| 2147 | 2497 | 1.006868 | 0.240093 | 0.334529 | 0.339180 | 0.335822 | 0.275595 | 0.062141 | 0.324242 | 0.275030 |
| 2148 | 2498 | 1.007111 | 0.240608 | 0.335397 | 0.340243 | 0.336710 | 0.276414 | 0.062232 | 0.325424 | 0.276053 |
| 2149 | 2499 | 1.007263 | 0.241110 | 0.336293 | 0.341387 | 0.337602 | 0.277276 | 0.062312 | 0.326167 | 0.276782 |
| 2150 | 2500 | 1.007191 | 0.241731 | 0.337202 | 0.342628 | 0.338453 | 0.278245 | 0.062381 | 0.326506 | 0.277072 |
2151 rows × 10 columns
The USGS spectral library contains many more spectra provided as csv files. We allow you to import a folder of them.
SpectralLibrary.import_USGS (directory:str, sort:bool=False)
Import the ASD files from directory with optional names sorted.
SpectralLibrary.import_USGS (directory:str, sort:bool=False)
Import the ASD files from directory with optional names sorted.
usgs_path = "ASCIIdata_splib07b/ChapterV_Vegetation/"
sl = SpectralLibrary()
sl.import_USGS(usgs_path)UserWarning: Spectral Library file required! Please re-initialised with the file.
sl = SpectralLibrary()Added folder of ASD spectra to spectral libraryThe plotted spectra have all be interpolated to remove all the NaNs.
Here we plot all the ASD data in the USGS Splib07b Chapter V Vegetation database.
It is not easy to visualise all of the USGS spectra in one plot. There are so many that the legend decided to pack up and go home. The one spectra with the sharp rise near 1000 nm is from a spectra that did not include any information in the visible region which messed with the interpolation. If you are only interested in spectral matching in the infrared, this is not a problem. Likewise, if you are interested in spectral matching in the visible, then this spectra shouldn’t be detected close to anything real.
Used later for interactive empirical line calibration. Since OpenHSI camera data can be processed to radiance, we use the 6SV radiance to guess what the spectral library will be in radiance. Then we compare radiances to calculate what known target to use in the ELC algorithm and hence find reflectance.
SpectralMatcher (cal_path:str, speclib_path:str=None)
Match OpenHSI spectra against spectral library using Spectral Angle Mapper algorithm. Requires calibration pkl file with the saved 6SV model.
| Type | Default | Details | |
|---|---|---|---|
| cal_path | str | path to spectral library (pandas DataFrame) | |
| speclib_path | str | None |
SpectralMatcher.topk_spectra (spectrum:<built-infunctionarray>, k:int=5, refine=True)
Match a spectrum against a spectral library spectra. Return the top k. Set refine to find closest match using mean squared error on the top k results.
| Type | Default | Details | |
|---|---|---|---|
| spectrum | array | spectrum array to compare against the loaded spectral library | |
| k | int | 5 | number of top matches to return |
| refine | bool | True | minimise the amplitude differences once similar spectra are found |
| Returns | DataFrame | results sorted from best match to worst |
SpectralMatcher.topk_spectra (spectrum:<built-infunctionarray>, k:int=5, refine=True)
Match a spectrum against a spectral library spectra. Return the top k. Set refine to find closest match using mean squared error on the top k results.
You can use this code snippet to find the top k=5 spectral matches. The Spectral Angle Mapper will say all gray calibration tarps have very similar spectra (spectrally flat) regardless of their percentage reflectance. The refine is there to refine the closest matches by mean squared error (which is not as fast).
sm = SpectralMatcher(cal_path="../assets/cam_calibration.nc",speclib_path="../assets/speclib.pkl")
topk = sm.topk_spectra( some_radiance_array, k=5, refine=True)
print(topk)topk is a pd.DataFrame with the first column as the labels in the spectral library. The second column is the score between [-1,+1] with better matches having score almost at 1.0.
You can visualise the matches with
and the transparency level depends on the score so poorer matches are faded out.
Essentially, Empirical Line Calibration (ELC) aims to solve a system of linear equations \[ L_i(\lambda) = a(\lambda)\rho_i(\lambda) + b(\lambda) \] where the radiance \(L\) is from what we measure, \(\rho\) is the known reflectance, and \(a\) and \(b\) are the multiplier and offset respectively. This is calculated for multiple pixels \(i\) and wavelengths \(\lambda\).
The implementation here involves solving these equations using linear algebra methods to find a least squares solution. It is fast enough to use this tool interactively.
Only load datacubes that have already been corrected to radiance to this interactive widget.
ELC (nc_path:str, old_style:bool=False, speclib_path:str=None)
Apply ELC for radiance datacubes
ELC.setup_callbacks ()
Setup the mouseover crosshair, and mouseclick callbacks.
This interactive tool requires a Python backend. There is no interactivity in the documentation site.
The top panel consists of an RGB view of your dataset using a robust visualisation technique to avoid outliers blowing out the contrast. When you mouseover the RGB view, a crosshair appears. There is a toolbar on the bottom left of the RGB view. From left to right, there are the: move tool, area select tool, scroll zoom tool, tap tool, box draw tool, figure save tool (requires additional packages to be installed), a reset tool, and a Bokeh logo.
Make sure you have the tap tool selected when viewing spectra so you don’t accidentally draw boxes.
To select the bright and dark targets for the ELC algorithm, select the box draw tool and double tap to start drawing (double tap again to finish drawing a box). All the pixels within the bounding boxes are used and updated immediately. A limit of 5 boxes are allowed. To delete drawn boxes, single click on a box and hold. While you holding, press the backspace key.
The bottom panel consists of a Reflectance and Radiance tab that you can switch between. When you click on a pixel on the RGB view, the spectra will update. If you are in the Reflectance tab, you will see the ELC and 6SV estimate alongside with the closest spectral matches in your spectral library. On the other hand, if you are in the Radiance tab, you will see the spectra labelled as “tap point” alongside estimates of the radiance (using the 6SV model) of the closest matches in your spectral library. You can tap on the legend to select or fade certain curves in the plot.
Finally, on the very bottom, there are two buttons to export the ELC or 6SV datacubes to NetCDF format (keeping the original capture times, camera temperatures if there are any, and metadata). These will save to the directory the original radiance datacube is loaded from.
Let’s see what the multiplier \(a(\lambda)\) and offset \(b(\lambda)\) was obtained using the gray and charcoal tarps as the bright and dark target.
a_plot = hv.Curve( zip(np.arange(len(elc.a_ELC)),elc.a_ELC), label="multiplier").opts(
xlabel="spectral band",ylabel="value")
b_plot = hv.Curve( zip(np.arange(len(elc.b_ELC)),elc.b_ELC), label="offset").opts(
xlabel="spectral band",ylabel="value")
a_plot * b_plotThe multiplier looks really similar to the 6SV estimate of the radiance which is not a surprise. Now to plot the results for all the calibration tarps.
df = pd.read_pickle("../assets/openhsi_radiance_select_targets.pkl")
df.plot(x='wavelength (nm)', y=df.columns[1:],figsize=(8,6),
xlabel="wavelength (nm)",ylabel="radiance (uW/cm^2/sr/nm)",
ylim=(0,None),title="Calibration targets").legend(loc="upper right",fontsize='xx-small')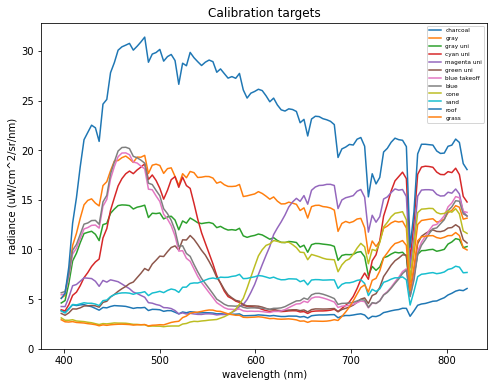
For exploring your datacubes.
DataCubeViewer (nc_path:str=None, old_style:bool=False, img_aspect_ratio:float=0.25, box_sz:tuple=(1, 1), **kwargs)
*Explore datacubes Optional key/val pair arguement overrides (**kwargs) ylim ylabel*
| Type | Default | Details | |
|---|---|---|---|
| nc_path | str | None | path to the NetCDF file |
| old_style | bool | False | if datacube is stored as cross-track, along-track, wavelength coords |
| img_aspect_ratio | float | 0.25 | aspect ratio for the datacube viewer |
| box_sz | tuple | (1, 1) | Any binning (nrows, ncols) around the tap point. Default is a single pixel |
| kwargs | VAR_KEYWORD |
DataCubeViewer.setup_callbacks ()
Setup the mouseover crosshair, and mouseclick callbacks.
This interactive tool requires a Python backend. There is no interactivity in the documentation site.